Research Article
Molecular Identification of Pathogens Causing Root Rot of Camellia oleifera in Tropical 
2 Institute of Agro-Environment and Soil of Hainan Academy of Agricultural Sciences, Hainan Key Laboratory of Arable Land Conservation, Haikou, 571100, P.R. China
 Author
Author  Correspondence author
Correspondence author
Molecular Pathogens, 2021, Vol. 12, No. 2 doi: 10.5376/mp.2021.12.0002
Received: 16 Nov., 2021 Accepted: 19 Nov., 2021 Published: 25 Nov., 2021
Zhao Z.X., Yan W.R., Xiao M., Xiao T.B., and Lei F., 2021, Molecular identification of pathogens causing root rot of Camellia oleifera in tropical, Molecular Pathogens, 12(2): 1-10 (doi:10.5376/mp.2021.12.0002)
Root rot of Camellia oleifera is a common root disease which has become one of the main diseases affecting the production of Camellia Oleifera oil in Hainan. In order to understand the pathogenic species and their diversity, and then to find out effective prevention strategies, increase the rate of forestation in Hainan, the root rot of the pathogen was initiated by the methods of morphological analysis, molecular identification and Phylogenetic analysis Root rot samples were collected from Yunlong, Wuzhi Mountain, Tongshi, Hongshan, Qiongzhong, Baisha, Wenchang, and so on. The pathogen of Camellia oleifera root rot disease was studied by routine tissue separation and purification, morphological identification, DNA sequence determination and analysis of ITS. At the same time, methods of comparing Blast in Genbank database, building the system evolutionary tree, and phylogenetic analysis were used in this research. Morphological identification, molecular identification, and subsequent phylogenetic analysis showed that there were two pathogens of Camellia Oleifera root rot in Hainan, one was Fusarium oxysporum, the other was Fusarium proliferatum. And Fusarium proliferatum was the major pathogenic type. Morphology on the representative strains could be divided into two classes, purple pigment and brown pigment. Sequence alignment and phylogenetic analysis found that the purple pigment representative strains were 100% sequence similarity with Fusarium proliferatum, and get together on the same branch; Brown pigment representative strains were 100% sequence similarity with Fusarium oxysporum, and get together on the same branch. This is the first report of Camellia Oleifera root rot caused by two or more pathogens alone or combined infection. The results of this study will be helpful to the breeding of Camellia Oleifera resistance and the research of its pathogenic mechanism.
Camellia oleifera, also known as camellia tree and camellia oil tree, is a small evergreen tree of the genus Camellia in Theaceae. Its name comes from the fact that its seeds can be pressed to produce camellia oil. There are thousands of years of planting history in China (He, 2010, Modern Economic Information, (10): 158, 187). It is mainly distributed in Hunan, Jiangxi, Hainan, Guizhou, Yunnan, Zhejiang, Guangdong, Guangxi, Chongqing, Anhui and other southern provinces (Zheng et al., 2016). Camellia oleifera in Hainan has a long history of native distribution, most of which are in wild or semi-wild state (Chen et al., 2017, Tropical Forestry, 45(1): 49-52). In recent years, the Hainan provincial government and some cities and counties have successively issued a number of policies on the development of Camellia oleifera, and incorporated it into the development plan. By 2017, the planting area of Camellia oleifera reached 5.63×103 hm2, mainly in Haikou, Qionghai, Chengmai, Ding'an, Tunchang, Lingao, Qiongzhong, Wuzhishan, Danzhou and other more than 10 cities and counties (Dai et al., 2017, Tropical Agricultural Engineering, 41(4): 61-64). However, with the intensification of planting, scale, deterioration of ecological environment and uneven seedlings in the base, the seedlings and young forest stage of Camellia oleifera in Hainan are facing the test of high incidence of root rot. From 2017 to 2019, it was found that root rot disease increased year by year, with the incidence of 50% in seedling stage, 10% to 20% in young forest stage, and more than 30% in severe cases, which has become one of the main diseases affecting Camellia oleifera in Hainan.
Camellia oleifera root rot disease is a devastating soil-borne disease in seedling stage. In the early stage, the roots of the affected seedlings turn brown to dark black, and then the root epidermis decayed and fell off, extending to the junction of the roots and stems on the ground, affecting water and nutrient transport, obstructing the growth of seedlings, stunting the plants, turning yellow and falling off the leaves, and even the whole plant dying, seriously affecting the forest and subsequent yield of Camellia oleifera. In order to effectively control the development of root rot in seedling stage, it is crucial to know the pathogen species. However, there are different reports on its pathogenic bacteria. Huang and Xiao (2015, Agricultural Science-Technology and Information, (12): 26-28) believed that the pathogen of Camellia oleifera root rot was Sclerotium rolosii. Peng (1998, Hunan Forestry Science & Technology, 25(4): 25-27) and Cao et al. (1993, Proceedings of the National Symposium on Promoting Forest Science and Technology, pp.323-326) reported that the pathogen was Cylindrocarpon destruclans. However, Li et al. (2008), Song et al. (2010), Hao (2009) and Liu (2011) believed that the pathogen of Camellia oleiferum root rot was Fusarium proliferatum, which had all the characters and morphological characteristics of Fusarium proliferatum. Ma et al. (2019, Anhui Agricultural Sciences, 47(5): 154-156) believed that the pathogen of Camellia oleifera root rot was Fusarium sp. Fungi, but did not specify any subspecies. At present, the pathogen of Hainan camellia root rot is not clear. Based on this, this study carried out morphological and molecular identification and evolutionary development analysis on the pathogenic bacteria of Hainan Camellia oleifera root rot disease, which laid a research foundation for preventing the disease and improving the forestation rate of Hainan Camellia oleifera.
1 Results and Analysis
1.1 Sample collection, isolation and pathogen verification
Seventy-eight samples of root rot were collected, and 47 pure pathogenic strains were obtained by tissue isolation, PDA plate culture, secondary transfer, and single spore isolation. HS-18 and T2FS-23 were inoculated in Camellia oleifera seedlings, and root rot appeared 20 d later, and the control grew well. The isolates which were consistent with the morphology of colonies and spores before inoculation could be isolated from the diseased seedling roots.
1.2 Morphological identification
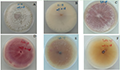
A total of 47 monocellular strains were isolated. According to the color of the back of the plate cultured on PDA medium for 7 days, they could be divided into three categories, producing purplish red pigment, tawny pigment and colorless pigment (Figure 1). The aerial mycelia of white cotton flocculent were rapidly overgrown on PDA medium, and the colony was regular and round. The culture medium at the bottom of the colony did not change color at the early stage, and gradually turned purple, purplish red or tawny at the later stage, and there were ringlets formed. On further observation, it was found that the bottom of the culture showed a colorless plate at 7 days, and with the extension of culture time (more than 15 d) (Figure 1A; Figure 1B), its bottom gradually shows mauve and purplish red.
|
Figure 1 Colony morphology of pathogenic Fungi Note: A, B: Positive and negative form of HS-18; C, D: Positive and negative form of T2FS-29; E, F: Positive and negative form of T2FS-23 |
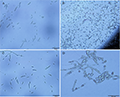
T2FS-23, HS-18 and T2FS-29 showed two types of conidia (Figure 2A; Figure 2B; Figure 2C). Many microconidia, nearly round, elliptic, ovoid, some slightly curved, single cell with 1 or 2 septa, size of 1~2.5 μm×5~12.2 μm; Macroconidia sickle-shaped, slightly pointed at both ends, with 3~5 septa, less in number, size of 3.3~6.5 μm ×30~35.5 μm; A small number of chlamydospore occur singly in mycelia and are spherical or subspherical (Figure 2D). In addition, the number of macroconidia of T2FS-23 and HS-18 was significantly higher than that of T2FS-29, and the sickle-shaped features of both ends were obvious. T2FS-29 was not significant. This may be caused by the inconsistent growth rate and sporulation rate of the strain on the medium plate. In conclusion, based on the identification methods of plant pathogen mycology (Lu, 1997, China Agriculture Press, pp.170-175), it was preliminarily concluded that the pathogen type of Hainan camellia root rot may be Fusasrium spp., according to the morphological characteristics of colonies and spores.
|
Figure 2 Conidia and chlamydospore morphology of pathogen Note: A: Macroconidia and Microconidia of T2FS-23; B: Macroconidia and Microconidia of T2FS-29; C: Macroconidia and Microconidia of HS-18; D: Chlamydospore |
1.3 Genomic DNA extraction of Camellia oleifera root rot fungus
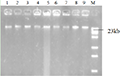
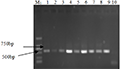
After extraction and purification, the DNA bands on the electrophoresis diagram (Figure 3) were all about 23 kb. The DNA concentration was estimated to be between 100 and 300 ng/μL according to the brightness of the bands. Uv spectrophotometer tests found that the OD260/OD280 values of T2FS-23, HS-18, T2FS-29, TS-01, TS-05, TS-09, YL-11, QZ-1 and WC-18 were 1.82, 1.75, 1.84, 1.77, 1.71, 1.83, 1.74, 1.87 and 1.73, respectively. It shows that the purity is relatively high, and the pollution of protein or other impurities to the sample is small. The concentration and purity of the DNA template can meet the requirements of PCR reaction.
|
Figure 3 Genomic DNA of some pathogenic strains Note: M: λ DNA/HindIII; 1~9: T2FS-23, HS-18, T2FS-29, TS-01, TS-05, TS-09, YL-11, QZ-1 and WC-18 |
1.4 PCR amplification and sample sequencing
The bands of the amplified products were detected by 1% agarose gel electrophoresis and ranged from 500 bp to 600 bp (Figure 4). Recycle with gel recovery kit (produced by Beijing TransGen Biotech). 50 μL EB was dissolved and 5 μL was detected by 1.0% agarose gel electrophoresis. The rest was directly sent to Beijing Liuhe BGI Technology Co., Ltd. for sequencing. Remove the sequencing results in NCBI first carrier sequence interface (www.ncbi.nlm.nih.gov/tools/vecscreen/), remove excess carrier sequence, and then after a BLAST homology comparison, to determine taxonomic status of Camellia oleifera root rot fungi in Hainan.
|
Figure 4 ITS gene on Strains of Camellia Oleifera root rot was amplified by PCR Note: M1, M2: DNA Marker Trans2k plus; 1~10: T2FS-23, HS-18, T2FS-29, TS-01, TS-05, TS-09, YL-11, QZ-1, WC-18 and control of water |
1.5 Sequence analysis
After removing redundant carrier sequences, two kinds of sequences are obtained. The total length of one base was 519 bp, and the similarity was 100%, and the representative strains are HS-18, T2FS-29, TS-01, TS-09, YL-11, QZ-1 and WC-18 strains, that is, the strain produced purplish red and colorless at the bottom of the medium. One with a total length of 545 bp and 100% similarity to each other, with the representative strains of TS-05 and T2FS-23, and produced tawny pigment at the bottom of the medium. Therefore, this is basically consistent with the results of morphological classification.
Blast comparison of ITS sequences of T2FS-29 in Genbank database (Figure 5) showed that a large number of ITS sequences of the same genus and different species with high homology were obtained. Among them, Fusarium proliferatum isolate RS_79 (Genbank N.: MK332493), Fusarium proliferatum strain CanR-8 (Genbank N.: JF817300), Fusarium isolate 165PG/F (Genbank N.: GU066714), Fusarium proliferatum isolate CLFpr2 (Genbank N.: MN481204), Fusarium proliferatum strain D2 (Genbank N.: EU151485), Fusarium proliferatum isolate ZXY5 (Genbank N.: MK158221), Fusarium proliferatum isolate F039 (Genbank N.: KU891521), Fusarium proliferatum strain E-2018 (Genbank N.: MK372368) had the highest similarity, all of which were over 99%, and the base sequences were different only at sites 335, 499 and 510, where the bases of T2FS-29 were C, C and C, respectively. The bases of Fusarium proliferatum isolate RS_79 were T, T and G, respectively. Landeweert et al. (2003) studied that fungi could be assigned to the same species if the sequence similarity of ITS region reached 99% or more. When the sequence similarity is between 95% and 99%, it can be considered as the same genus. If the sequence similarity is less than 95%, it can be considered as the same family based on morphological observation. According to the results, strains T2FS-29, HS-18, TS-01, TS-09, YL-11, QZ-1 and WC-18 could be preliminarily identified as Fusarium proliferatum.
|
Figure 5 Comparison of T2FS-29 and RS_79 (Genbank N. MK332493) ITS sequences Note: gb/MK332493/ Fusarium proliferatum; Length=558, Score=942 bits(510), Expect=0.0, Identities=516/519 (99%), Gaps=0/519(0%), Strand=Plus/Plus; Query: T2FS-29, Sbjct: MK332493 |
Similarly, a large number of ITS sequences belonging to different species with high homology were obtained by Blast comparison search of T2FS-23 ITS sequences in Genbank database. Fusarium oxysporum isolate FOM-12 (Genbank N.: MN272279), Fusarium oxysporum isolate FOB-29 (Genbank N.: MN272279), Fusarium oxysporum f.sp.ciceris (Genbank N.: KU097320), Fusarium oxysporum strain MJ-23 (Genbank N.: KF751873), Fusarium oxysporum (Genbank N.: JN400690), Fusarium oxysporum isolate B10 (Genbank N.: MN567668), Fusarium oxysporum f.sp.ciceris strain FOC19 (Genbank N.: KF534747) had the highest similarity (99.82%), except for the difference in the sequence of the base at site 273. The base of T2FS-23 was G, while that of Fusarium oxysporum isolate FOM-12 was A (Figure 6). And the similarity of Fusarium proliferatum (Genbank N.: LS422789), Fusarium proliferatum strain FP-4 (Genbank N.: MK814399), Fusarium proliferatum isolate FMB-CP1-FR (Genbank N.: MK583509) and Fusarium proliferatum strain Z4 (Genbank N.: MH486974) was 98.90%. Therefore, according to the classification and comparison of Landeweert et al. (2003), T2FS-23 and TS-05 can be preliminarily identified as Fusarium oxysporum.
|
Figure 6 Comparison of T2FS-23 and Fusarium oxysporum isolate FOM-12(MN272279)ITS sequences Note: gb/MN272279/ Fusarium oxysporum: Length=568, Score=1002 bits(542), Expect=0.0, Identities=544/545 (99%), Gaps=0/545(0%) Strand=Plus/Plus; Query: T2FS-23, Sbjct: MN272279 |
1.6 Phylogenetic analysis of HS-18 and T2FS-29 strains
HS-18 and T2FS-29 ITS sequences were compared with BLAST in NCBI, and several ITS sequences with high similarity were obtained, all of which were Fusarium populations, with the highest similarity of 99.62% in Fusarium proliferatum. Fusarium verticillioides strain CBS (Genbank N.: MH864460) and Gibberella moniliformis (Genbank N. HQ176445) followed, and the similarity was over 98%. Except for source reference sequences, the similarity with Fusarium fujikuroi isolate TB2-3P2 (KX683427) was low, only 97%.
HS-18 and T2FS-29 clustered on a large dendritic branch with multiple sequences of Fusarium proliferatum isolate RS_79 (Genbank N.:MK332493), Fusarium proliferatum strain CanR-8 (Genbank N.: JF817300), Fusarium proliferatum isolate 165PG/F (Genbank N.: GU066714), Fusarium proliferatum isolate CLFpr2 (Genbank N.: MN481204), Fusarium proliferatum strain D2 (Genbank N.: EU151485), Fusarium proliferatum isolate ZXY5 (Genbank N.: MK158221), Fusarium proliferatum isolate F039 (Genbank N.: KU891521) and Fusarium proliferatum strain E-2018 (Genbank N.: MK372368) cluster on the same and narrower branch (Figure 7). It was confirmed that HS-18 and T2FS-29 were Fusarium proliferatum based on phylogenetic tree rules and morphological characteristics. This is consistent with the reports of Hao (2009) and Liu (2011) on the pathogenic bacteria of Camellia oleifolia root rot in Hunan and Hubei.
|
Figure 7 Phylogenetic tree of ITS sequence analysis based on HS-18 and T2FS-29 |
1.7 Phylogenetic analysis of representative strains TS-05 and T2FS-23
After BLAST comparison of TS-05 and T2FS-23 ITS sequences in NCBI, several ITS sequences with high similarity were obtained, which were also Fusarium populations, and Fusarium oxysporum had the highest similarity (99.82%). It was 98.90% similar to Fusarium proliferatum (Genbank N.: LS422789) and Fusarium proliferatum strain FP-4 (Genbank N.: MK814399). The similarty with Fusarium verticillioides strain JG32-2 (Genbank N.: KJ699107) and Gibberella moniliformis strain FJAT-3756 (Genbank N.: JQ277275), Gibberella moniliformis isolate 840 (Genbank N.: JN232130), Gibberella moniliformis isolate 819 (Genbank N.: JN232125) was lower (more than 97%). Except for the source reference sequence, the similarity with Fusarium dlaminii strain CBS738 (Genbank N.: MH862668) was the lowest, only 95%.
TS-05 and T2FS-23 were clustered on a large dendritic branch with multiple sequences of Fusarium, while they were clustered on the same smaller branch (Figure 8) with Fusarium oxysporum isolate FOM-12 (Genbank N.: MN272279), Fusarium oxysporum isolate FOB-29 (Genbank N.: MN272278), Fusarium oxysporum f.sp.ciceris (Genbank N.: KU097320), Fusarium oxysporum strain MJ-23 (Genbank N.: KF751873), Fusarium oxysporum (Genbank N.: JN400690), Fusarium oxysporum isolate B10 (Genbank N.: MN567668) and Fusarium oxysporum f.sp.ciceris (Genbank N.:KF534747). According to the method of phylogenetic tree identification and morphological characteristics, TS-05, T2FS-23 and other pathogens were identified as Fusarium oxysporum. This is different from the reports of Hao (2009) and Liu (2011) on the pathogenic bacteria of Camellia oleifolia root rot in Hunan and Hubei. Combined with the observation results of pathogen morphological characteristics, it could be concluded that the root rot of Hainan Camellia oleifera was caused by the compound infection of Fusarium proliferatum and Fusarium oxysporum, some of which are mainly Fusarium oxysporum and some are Fusarium proliferatum. Fusarium oxysporum and Fusarium proliferatum could be isolated in most places.
|
Figure 8 Phylogenetic tree of ITS sequence analysis based on TS-05 and T2FS-23 |
2 Discussion
Morphological characteristics, supplemented by molecular biology, have been an effective way to identify pathogenic microorganisms in recent years (Zhao et al., 2019; Liu, 2011; Sixto et al., 2018). In this study, the pathogen species of Camellia oleifera root rot in Yunlong, Wenchang, Wuzhishan, Hongshan, Tongshi and Qiongzhong were systematically studied by means of field symptom observation, morphological observation, ITS sequence determination and sequence analysis. The symptoms and occurrence of root rot of Hainan Camellia oleifera were clarified. To clarify the occurrence regularity of Hainan Camellia oleifera root rot; Two kinds of pathogen were identified against root rot of Hainan Camellia oleifera, one was Fusarium proliferatum and the other was Fusarium oxysporum. On the morphological observation, two different types of pigments (purplish red and tawny) were produced on the plate after 7 d of culture (including above), and the molecular identification and phylogenetic development analysis just agreed with the morphological observation results, indicating that there were two pathogens of Fusarium proliferatum and Fusarium oxysporum. It was also the first time that the root rot of Camellia oleifera in Hainan was reported to be caused by the compound infection of Fusarium proliferatum and Fusarium oxysporum, some of which are mainly Fusarium oxysporum and some are Fusarium proliferatum. Fusarium oxysporum and Fusarium proliferatum could be isolated in most places. This study provided scientific basis for further study on the epidemic law of Camellia oleifera root rot in Hainan and disease resistance breeding.
3 Materials and Methods
3.1 Collection and isolation of Camellia oleifera root rot samples
Seventy-eight Camellia oleifera samples were collected from Yunlong, Wuzhishan, Tongshi, Hongshan, Qiongzhong, Baisha and Wenchang of Haikou. Forty-seven pure pathogenic strains were isolated (Table 1). Conventional tissue separation method (Fang, 2003, China Agriculture Press, pp.122-124) was used for isolation of pathogen. That is, the junction of rotten roots and stems was taken, the epidermis of rotten roots was removed, and the tissues at the junction of xylem and phloem were cut into 2.5 mm×2.5 mm×2.5 mm pieces. The tissues were disinfected with 0.1% hg for 30 s, soaked in 70% alcohol for 30 s, washed with sterile water, and dried in air. Finally, the tissues were transferred to PDA plate and cultured upside down in a temperature box at 25℃. Referring to the research method of Zhao et al. (2017), the obtained colonies were isolated and numbered. They were cultured on a PDA inclined plane and stored in a refrigerator at 4℃ for future use.
|
Table 1 Pathogenic strains of Camellia Oleifera root rot in Hainan |
3.2 Verification of pathogenic bacteria
HS-18 and T2FS-23 were selected for pathogen verification based on phenotypic differences on plate. HS-18 and T2FS-23 were inoculated in PD liquid medium at 28℃, 180 r/min, shaking culture for 48 h, sterile double gauze filtration, 105~107 cfu spore suspension, irrigating method inoculated in Camellia oleiferia seedlings treated with prior root injury, 10 mL/plant, 3 replicates, and water control was set up. After the onset of disease, samples were taken and isolated to compare whether the pathogens were consistent before and after inoculation.
3.3 Morphological characteristics of pathogen
The pathogen was inoculated on PDA medium at 28℃ for 14 d. The morphology of plate colony (including mycelium density, color of back, etc.) was observed. According to The Chinese Crop Diseases and Insect Pests (Edition 3) Volume (Guo, 2015, China Agriculture Press, pp.182-184), microscopic examination was performed to compare the morphology, size and type of conidia and preliminarily determine the type of pathogenic bacteria.
3.4 Genomic DNA extraction, PCR amplification and sequencing
The pathogen was cultured on a PDA tablet, scrape the hyphae, the genomic DNA of Camellia oleifera root rot bacteria was extracted and purified by improved CTAB-lysozyme -protease K method. Referring to the research method of Zhao et al. (2013), the ITS sequences of representative strains were amplified by PCR. A small amount of amplified products were detected by electrophoresis and sent to Beijing Liuhe BGI for sequencing.
3.5 Sequence and phylogenetic analysis
The ITS sequences were removed online from NCBI and BLAST nucleotide alignment was performed. Meanwhile, the ITS sequences of the same species with high similarity were downloaded, and Rhizoctonia solani strain 5 (Genbank N.: MK294853) as exogenous reference, Clustal W bioinformatics software was used to align the sequences, and the proximity method in MEGA X (Kumar et al., 2018) bioinformatics software was used to construct phylogenetic tree and analyze the phylogenetic development relationship of pathogens.
Authors’ contributions
ZZX is the experimental designer and executor of this study. ZZX completed data analysis and wrote the first draft of the paper; YWR, XM, LF participated in experimental design and analysis of experimental results; XTB was the architect and principal of the project, guiding experimental design, data analysis, paper writing and modification. All authors read and approved the final manuscript.
Acknowledgments
This study was supported by the Key Research and Development Program of Hainan Province (ZDYF2018093) and the Financial Science and Technology Program of Hainan Province.
Hao F., 2009, The research on camellia oleifera root rot and its PCR rapid, Thesis for M.S., Life Science and Technology Institute, Central South University of Forestry and Teclmology, Supervisor: Zhou G.Y., pp26-27
Kumar S., Stecher G., Li M., Knyaz C., and Tamura K., 2018, MEGA X: molecular evolutionary genetics analysis across computing platforms, Mol. Biol. Evol., 35(6): 1547-1549
https://doi.org/10.1093/molbev/msy096
Li H., Zhou G.Y., and He M.J., 2008, Study on biological characteristics and molecular identification of pathogens causing root rot of Camellia oleifera, Xinan Linxueyuan Xuebao (Journal of Southwest Forestry College), 28(5): 45-48, 56
Liu S.B., 2011, A preliminary study of root rot pathogen on camellia, Thesis for M.S., Plant Science and Technology Institute, Huazhong Agricultural University, Supervisor: Hou M.S., pp.16-17
Landeweert R., Leeflang P., Kuyper T.W. Hoffland E., Rosling A., Wernars K., Smit E., 2003, RPL, THANW K, etal. Molecular identification of ectomycorrhizal mycelium in soil horizons, Applied and Envimmn ental Micmhiology, 69(1): 327-333
https://doi.org/10.1128/AEM.69.1.327-333.2003
Sixto V.F., José A.G., Sergio H.V., Carlos A.L., and Jesús E.M., 2018, Occurrence of Fusarium oxysporum causing wilt on pepper in Mexico, Canadian Journal of Plant Pathology, 40(2): 238-247
https://doi.org/10.1080/07060661.2017.1420693
Song G.T., Zhou G.Y., Liu J.A., Li H., and Li L., 2018, Isolation and characterization of antagonistic endophytic bacteria again Fusarium proliferatum, Zhiwu Baohu Xuebao (Acta Phytophylacica Snica), 37(2): 137-142
Zhao Z.X., Ling B., Yan W.R., Zeng X.P., Xiao M., and Chen Y., 2019, Isolation, identification and phylogenetic analysis of pepper wilt, Fenzi Zhiwu Yuzhong (Molecular Plant Breeding),17(19): 6383-6389
Zhao Y.Q., Yu H.R., Shi K., Zhang L.J., Zhang D.M., Zhu P.T., Cai C., and Jia A.M., 2017, Identification of the pathogen causing leaf spot on sorghum, Zhiwu Bingli Xuebao (Acta Phytopathologica Sinica), 47(2): 282-285
Zhao Z.X., Chen Y., Chen M.C., Fu M.Y., Wang S.Y., and Xiao T.B., 2013, Identification of dioscorea stem rot disease in Hainan, Jijinzuxue Yu Yingyong Shengwuxue (Genomics and Applied Biology), 32(6): 761-766
Zheng D.J., Pan X.Z., Zhang D.M., Xie L.S., Zeng J.H., Zhang Z.L., and Ye H., 2016, Survey and analysis on tea-oil camellia resource in Hainan, Xibei Linxueyuan Xuebao (Journal of Northwest Forestry Niversity), 31(1): 130-135, 169
. PDF(1991KB)
. HTML
Associated material
. Readers' comments
Other articles by authors
. Zhixiang Zhao
. Wanrong Yan
. Min Xiao
. Tongbin Xiao
. Fei Lei
Related articles
. Camellia oleifera
. Root rot disease
. Molecular identification
. Phylogenetic analysis
Tools
. Email to a friend
. Post a comment
.png)







